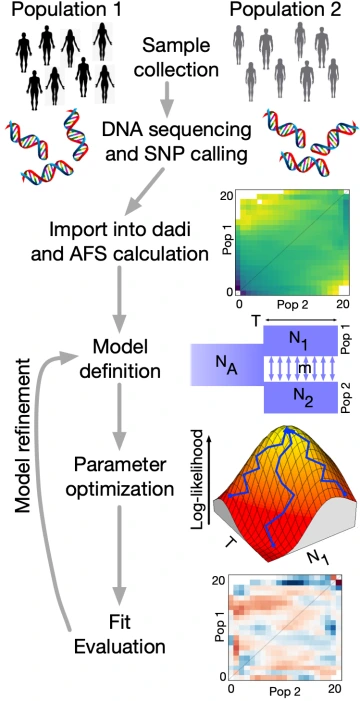
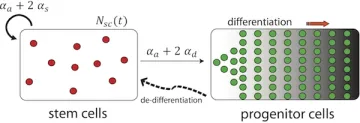
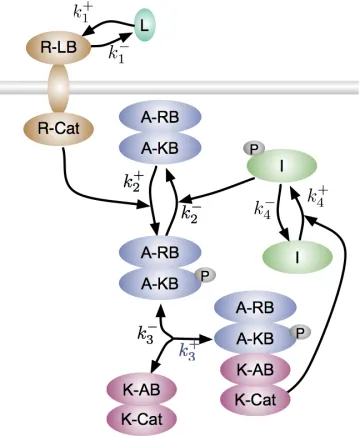
Cancer is fundamentally an evolutionary process, as tumor cells acquire mutations to enhance their growth, outcompete their neighbors, and evade the immune system. We thus apply concepts and tools from population genomics to understand tumor development.
Tumor genomic sequencing data are critical for understanding genetic variation within tumors, which affects their development and recurrence. In particular, low frequency mutations within tumors are strongly informative about tumor development, but they are difficult to identify among sequencing errors. To improve identification, we developed a Bayesian approach that incorporates the fact that every tumor has a unique spectrum of mutational types that it tends to generate (Mannakee and Gutenkunst NAR: Genomics and Bioinformatics 2020). High-frequency variants can be used to identify this spectrum, so it can be applied to improve identification of low-frequency variants, enabling more accurate evolutionary inferences.
A key goal of cancer evolution research, with applications to cancer prevention, is to understand the factors that hasten the acquisition of tumor driver mutations. Tumors can quickly acquire driver mutations by elevating their mutation rate, but not all tumors do this. In theoretical work, we established an alternative possibility, that tumors may benefit from altering not the total rate of mutations but rather the spectrum of types of mutations (Tuffaha et al. Am Naturalist 2023). Using data from thousands of tumors, we empirically validated this prediction (Tuffaha et al. Mol Biol Evol 2025). Our analysis also suggested that hypoxia drives particularly potent changes in mutation spectra, a prediction we are following up on.
Mutations within tumors can not only promote growth, they can also generate novel antigens that provoke an immune response. By contrasting genomic data from tumors in immunocompetent versus immunocompromised patients, we recently identified key factors that identify immunogenic mutations (Borden et al. submitted). We are leveraging these insights to better identify tumor vaccine targets, and we are developing models to better understand the immune system’s effect on early tumor development.